Abstract
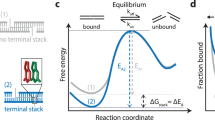
The specific hybridization of complementary sequences is an essential property of nucleic acids, enabling diverse biological and biotechnological reactions and functions. However, the specificity of nucleic acid hybridization is compromised for long strands, except near the melting temperature. Here, we analytically derived the thermodynamic properties of a hybridization probe that would enable near-optimal single-base discrimination and perform robustly across diverse temperature, salt and concentration conditions. We rationally designed ‘toehold exchange’ probes that approximate these properties, and comprehensively tested them against five different DNA targets and 55 spurious analogues with energetically representative single-base changes (replacements, deletions and insertions). These probes produced discrimination factors between 3 and 100+ (median, 26). Without retuning, our probes function robustly from 10 °C to 37 °C, from 1 mM Mg2+ to 47 mM Mg2+, and with nucleic acid concentrations from 1 nM to 5 µM. Experiments with RNA also showed effective single-base change discrimination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Saiki, R. K. et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491 (1988).
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Gunderson, K. L., Steemers, F. J., Lee, G., Mendoza, L. G. & Chee, M. S. A genome-wide scalable SNP genotyping assay using microarray technology. Nature Genet. 37, 549–554 (2005).
Koltai, H. & Weingarten-Baror, C. Specificity of DNA microarray hybridization: characterization, effectors, and approaches for data correction. Nucleic Acids Res. 36, 2395–2405 (2008).
DeLong, E. F., Wickham, G. S. & Pace, N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243, 1360–1363 (1989).
Amann, R. I., Krumholz, L. & Stahl, D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172, 762–770 (1990).
Seeman, N. C. Nanomaterials based on DNA. Annu. Rev. Biochem. 79, 65–87 (2010).
Rothemund, P. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Aldaye, F. A., Palmer, A. L. & Sleiman, H. F. Assembling materials with DNA as the guide. Science 321, 1795–1799 (2008).
Yin, P., Choi, H. M. T., Calvert, C. R. & Pierce, N. A. Programming biomolecular self-assembly pathways. Nature 451, 318–322 (2008).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand displacement reactions. Nature Chem. 3, 103–113 (2011).
Tyagi, S. & Kramer, F. R. Molecular beacons: probes that fluoresce upon hybridization. Nature Biotechnol. 14, 303–308 (1996).
Tyagi, S., Bratu, D. P. & Kramer, F. R. Multicolor molecular beacons for allele discrimination. Nature Biotechnol. 16, 49–53 (1998).
Tyagi, S. Imaging intracellular RNA distribution and dynamics in living cells. Nature Methods 6, 331–338 (2009).
Bonnet, G., Tyagi, S., Libchaber, A. & Kramer, F. R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA 96, 6171–6176 (1999).
Tsourkas, A., Behlke, M. A., Rose, S. D. & Bao, G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 31, 1319–1330 (2003).
Xiao, Y. et al. Fluorescence detection of single-nucleotide polymorphisms with a single, self-complementary, triple-stem DNA probe. Angew. Chem. Int. Ed. 48, 4354–4358 (2009).
Kolpashchikov, D. M. A binary DNA probe for highly specific nucleic acid recognition. J. Am. Chem. Soc. 128, 10625–10628 (2006).
Dave N. & Liu, J. Fast molecular beacon hybridization in organic solvents with improved target specificity. J. Phys. Chem. B 114, 15694–15699 (2010).
SantaLucia, J. & Hicks, D. The thermodynamics of DNA structural motifs. Ann. Rev. Biophys. Biomol. Struct. 33, 415–440 (2004).
Peyret, N. Prediction of Nucleic Acid Hybridization: Parameters and Algorithms. Doctoral thesis, Wayne State University (2000).
Tan, Z. J. & Chen, S. J. Nucleic acid helix stability: effects of salt concentration, cation valence and size, and chain length. Biophys. J. 90, 1175–1190 (2006).
Yurke, B., Turberfield, A. J., Mills, A. P., Simmel, F. C. & Neumann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Zhang, D. Y., Turberfield, A. J., Yurke, B. & Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007).
He, G., Rapireddy, S., Bahal, R., Sahu, B. & Ly, D. H. Strand invasion of extended, mixed-sequence B-DNA by γPNAs. J. Am. Chem. Soc. 131, 12088–12090 (2009).
Petersen, M. & Wengel, J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 21, 74–81 (2003).
Bommarito, S., Peyret, N. & SantaLucia, J. Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 28, 1929–1934 (2000).
Dirks, R. M., Bois, J. S., Schaeffer, J. M., Winfree, E. & Pierce, N. A. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 49, 65–88 (2007).
Zhang, D. Y. & Winfree, E. Robustness and modularity properties of a non-covalent DNA catalytic reaction. Nucleic Acids Res. 38, 4182–4197 (2010).
Temsamani, J., Kubert, M. & Agrawal, S. Sequence identity of the n–1 product of a synthetic oligonucleotide. Nucleic Acids Res. 23, 1841–1844 (1995).
Marras, S. A., Kramer, F. R. & Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 30, e122 (2002).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Li, Q., Luan, G., Guo, Q. & Liang, J. A new class of homogeneous nucleic acid probe based on specific displacement hybridization. Nucleic Acids Res. 30, e5 (2002).
Subramanian, H. K. K., Chakraborty, B., Sha, R. & Seeman, N. C. The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami. Nano Lett. 11, 910–913 (2010).
Gao, Y., Wolf, L. K. & Georgiadis, R. M. Secondary structure effects on DNA hybridization kinetics: a solution versus surface comparison. Nucleic Acids Res. 34, 3370–3377 (2006).
Kim, S. & Misra A. SNP genotyping: technologies and biomedical applications. Annu. Rev. Biomed. Eng. 9, 289–320 (2007).
Lizardi, P. M. et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet. 19, 225–232 (1998).
Isaacs, F. J., et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nature Biotechnol. 22, 841–847 (2004).
Venkataraman, S., Dirks, R. M., Ueda, C. T. & Pierce, N. Selective cell death mediated by small conditional RNAs. Proc. Natl Acad. Sci. USA 107, 16777–16782 (2010).
Acknowledgements
The authors thank M. Dai and P-S. Loh for assistance with mathematical analysis and J. Aliperti, E. Haney, R. Jungmann and T. Schaus for helpful suggestions during manuscript preparation. This work was funded by a Wyss Institute for Biologically Inspired Engineered faculty start-up fund, an NIH Director's New Innovator Award (1DP2OD007292), an NSF CAREER Award (CCF1054898) and an Office of Naval Research grant (N000141010827) to P.Y. D.Y.Z. is a Howard Hughes Medical Institute postdoctoral fellow, as part of the Life Sciences Research Foundation programme. There is a patent pending on the methods described in this work.
Author information
Authors and Affiliations
Contributions
D.Y.Z. conceived the project, designed and conducted the experiments, analysed the data and wrote the manuscript. S.X.C. conducted experiments, analysed the data and edited the manuscript. P.Y. conceived, designed and supervised the study, analysed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have a patent pending on the methods described in the manuscript.
Supplementary information
Supplementary information
Supplementary information (PDF 765 kb)
Rights and permissions
About this article
Cite this article
Zhang, D., Chen, S. & Yin, P. Optimizing the specificity of nucleic acid hybridization. Nature Chem 4, 208–214 (2012). https://doi.org/10.1038/nchem.1246
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1246
This article is cited by
-
Heteromultivalency enables enhanced detection of nucleic acid mutations
Nature Chemistry (2024)
-
Rapid, tunable, and multiplexed detection of RNA using convective array PCR
Communications Biology (2023)
-
Sensitive detection of SARS-CoV-2 on paper
Nature Biomedical Engineering (2022)
-
Photonic crystal enhanced fluorescence emission and blinking suppression for single quantum dot digital resolution biosensing
Nature Communications (2022)
-
Highly multiplexed rapid DNA detection with single-nucleotide specificity via convective PCR in a portable device
Nature Biomedical Engineering (2021)